Graduate Research
I. Deciphering the molecular maturation of mouse retinal ganglion cells
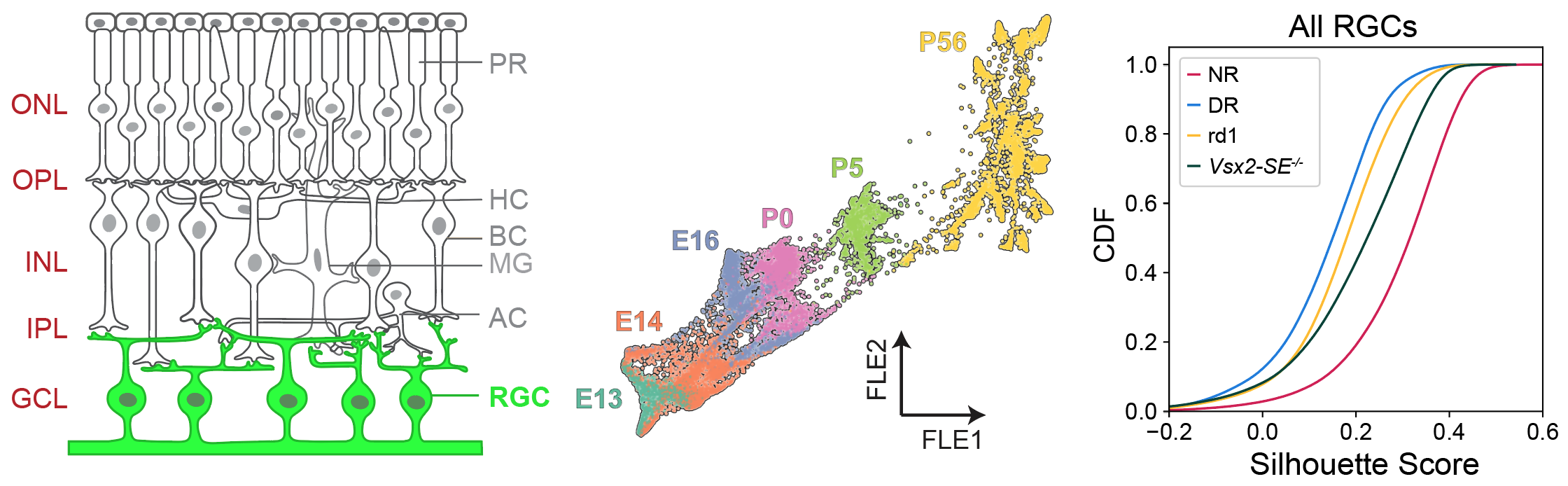
The development and connectivity of retinal ganglion cells (RGCs), the retina’s sole output neurons, are patterned by activity-independent transcriptional programs and activity-dependent remodeling. To inventory the molecular correlates of these influences, I applied high throughput single-cell RNA sequencing (scRNA-seq) to mouse RGCs at six embryonic and postnatal ages. I identified temporally regulated modules of genes that correlate with key phases of RGC development, ranging from differentiation and axon guidance to synaptic recognition and refinement. Next, I used these results as a foundation to analyze the transcriptomes of RGCs in mice lacking visual experience due either to dark rearing from birth or to genetic mutations that ablate bipolar or photoreceptor cells. 98.5% of visually deprived (VD) RGCs could be unequivocally assigned to a single RGC type based on their transcriptional profiles, suggesting that visual activity is dispensable for diversification of multipotential precursors into discrete types. However, visual deprivation significantly reduced the transcriptomic distinctions between RGCs, implying that activity influences RGC maturation or maintenance. These results provide a resource for mechanistic analyses of RGC differentiation and maturation, and for investigating the role of activity in these processes.
*=Equal contribution
- *Whitney, I., *Butrus, S., Sanes, J.R., Shekhar, K. Vision-dependent and -independent molecular maturation of mouse retinal ganglion cells. European Journal of Neuroscience (2023). In press.
II. Characterization of visual cortex plasticity at cell type resolution
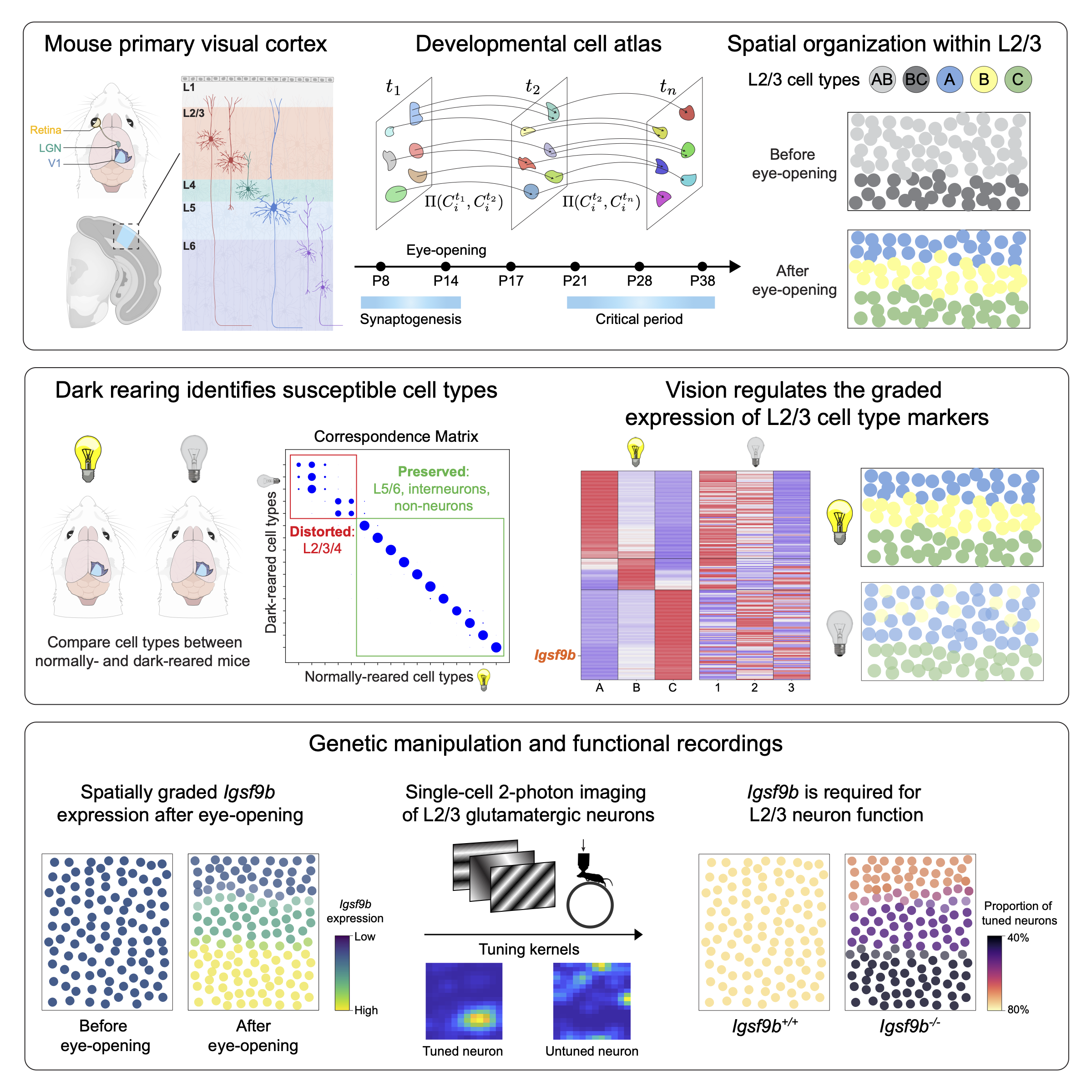
I began my PhD with an interest in combining molecular biology, experimental neuroscience, and machine learning to study visual cortex development at the resolution of its hundreds of cell types. I led a collaboration between our group and Profs. Larry Zipursky and Josh Trachtenberg’s groups at UCLA to study the development of visual cortex cell types as a function of sensory input. The role of postnatal experience in sculpting cortical circuitry, while long appreciated, is poorly understood at the level of cell types. We combined single-nucleus (sn) transcriptomics, statistical inference, sensory perturbations, genetics, and in vivo functional imaging to assemble a developmental transcriptomic atlas of postnatal mouse V1. Using this as a foundation, we discovered that: (1) vision is required for the establishment and maintenance of L2/3 glutamatergic types, but not other cell types in V1; (2) L2/3 glutamatergic cell types are organized as sublayers in V1 and form a transcriptomic continuum through the graded expression of ~200 genes; and (3) among these genes, Igsf9b, a vision-regulated cell adhesion molecule, is required in a graded fashion for the functional maturation of L2/3 glutamatergic neurons. In addition to challenging the prevailing hypothesis that genetically-hardwired mechanisms establish a static blueprint of cell types, our study establishes a framework for future investigations of how experience regulates neuronal organization in the brain.
*=Equal contribution
- *Cheng, S., *Butrus, S., *Tan, L., Sagireddy, S., Trachtenberg, J.T., Shekhar, K., Zipursky, L. Vision-dependent specification of cell types and function in the developing cortex. Cell (2022) 185, 2: pp. 311-327.
- Highlighted in Nature by Puiggros and Jabaudon (2022).
- Highlighted in UC Berkeley, College of Chemistry Press Release (2022).
- Highlighted in EurekAlert (2022).
- Butrus, S., Shekhar, K. Vision mediates molecular patterning in the developing visual cortex. Society for Neuroscience Meeting (2021).
- Butrus, S., Shekhar, K. Vision mediates molecular patterning in the developing visual cortex. UC Berkeley Center for Computational Biology Retreat (2021).
III. Python package for studying neuroprotection and axon regeneration using single-cell RNA sequencing
A major hallmark of the mammalian central nervous system is that certain populations of neurons exhibit far greater vulnerability to insults than others. The mechanisms underlying this selective neuronal vulnerability in the context of acute (e.g. traumatic injury) or chronic (e.g. neurodegenerative) disease have been difficult to dissect, but recent advances in single-cell RNA-seq (scRNA-seq) make it possible to compare patterns of gene expression among neuronal types that differ in vulnerability. As a postdoctoral fellow, my advisor Prof. Karthik Shekhar explored this strategy in the context of axotomy by analyzing the responses of mouse retinal ganglion cells (RGCs) to optic nerve crush (ONC), a well-studied model of traumatic injury. RGCs are a diverse class of projection neurons, with their diversity in mice comprising >40 discrete types, each with distinct morphological, physiological and molecular features. Using high-throughput scRNA-seq, he derived an atlas of 46 molecularly distinct RGC types. Following ONC, he found that ~85% of RGCs die within two weeks, and those that survive do not regenerate axons. To determine if specific types are lost at the same or different rates, he profiled RGCs using scRNA-seq from injured retinas at 0, 0.5, 1, 2, 4, 7, and 14 days post crush (dpc) and found cell type-specific axonal injury and regeneration patterns.
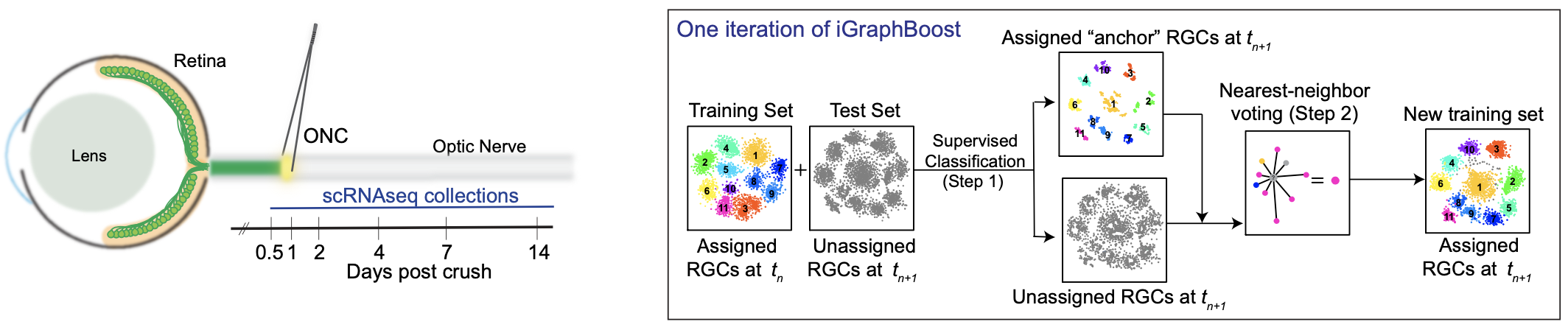
To generalize the computational methods used in this project to any study of neuronal vulnerability and make them accessible to researchers without an extensive background in machine learning, my colleague and I developed a code base in the programming language Python and wrote a step-by-step protocol describing its implementation. I described the computational framework used to analyze transcriptomic profiles of injured RGCs over the aforementioned time course, aiming to assess type-specific differences in survival, and identify gene signatures that correlate with, and may underlie, these differences. Following injury, extensive gene expression changes in RGCs make the inference of type identity challenging. To address this, I used an iterative supervised classification approach that leverages data from RGCs collected at the intermediate time points. In this approach, transcriptomic signatures of RGC types are successively redefined at each time to map the cells at the next time point. The underlying method, called iterative-GraphBoost (iGraphBoost), combines a two-step procedure involving gradient boosted trees (Step 1) and a graph-based voting scheme (Step 2). This dramatically increases cell type mapping efficiency at later time points and allows us to deconvolve gradual injury-related “state” changes from intrinsic type-specific gene expression programs.
- Butrus, S., Sagireddy, S., Shekhar, K. Defining selective neuronal resilience and identifying targets of neuroprotection and axon regeneration using single-cell RNA sequencing – computational approaches. Methods in Molecular Biology (2023). In press.
Undergraduate Research
I. Development and characterization of gold nanoparticles for plant genetic engineering
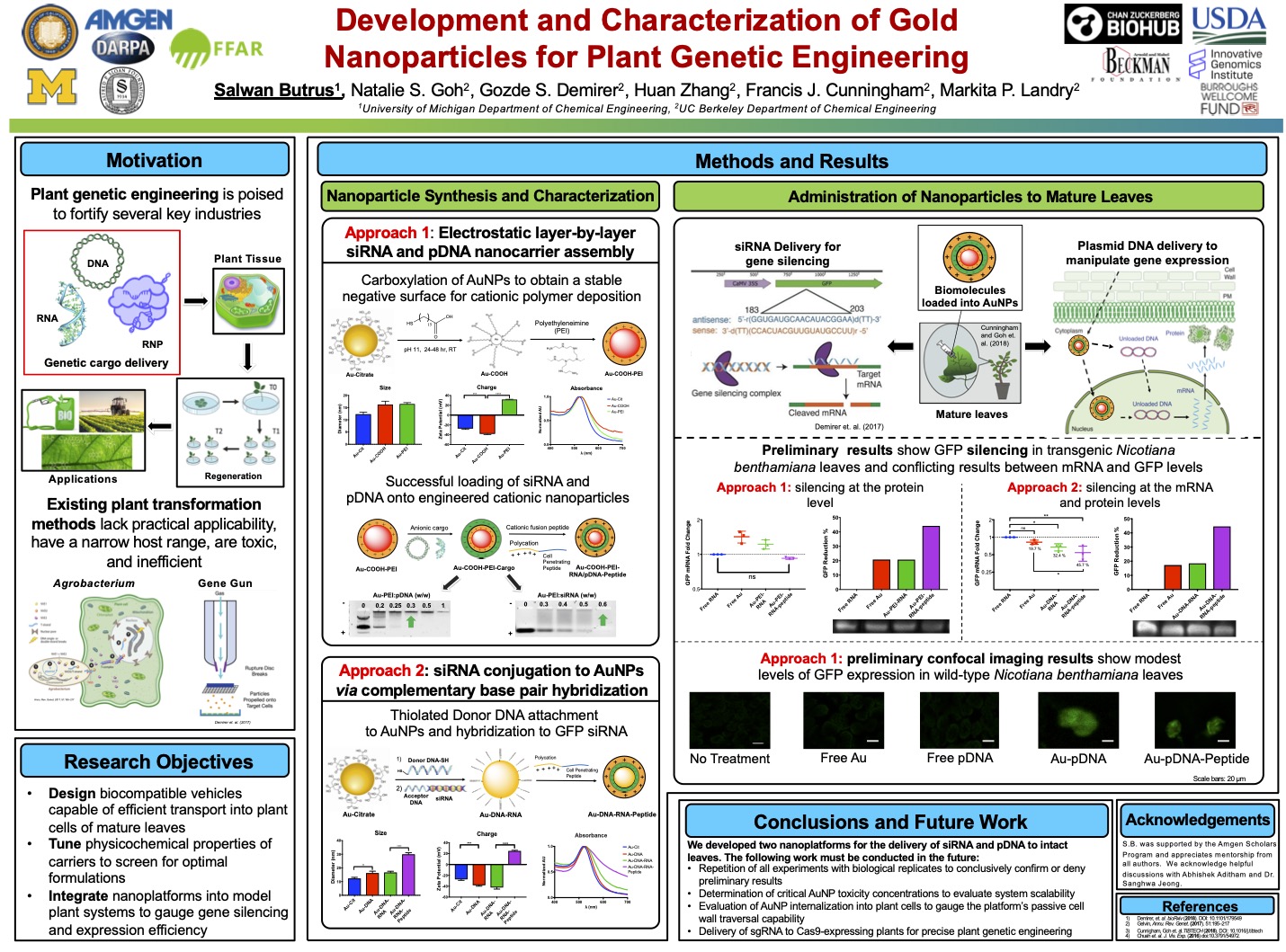
Plant genetic engineering is poised to strengthen the agricultural, pharmaceutical, and energy industries in the face of climate change and global population growth. However, traditional biomolecule delivery methods to plants are limited by low efficiency, narrow host range, toxicity, and limited practical applicability due to the transport challenge posed by the plant cell wall. I was intrigued by early work in using carbon nanotubes to transform mature plants in a non-toxic, passive, and species-independent manner that requires no transgene integration. I sought to gauge the promise of gold nanoparticles (AuNPs) as plant genetic engineering tools. By systematically tuning key physicochemical properties of the NPs to optimize bio-cargo loading efficiency, I developed the first two AuNP-based methods for delivering small interfering RNA and plasmid DNA to intact leaves for gene silencing and expression, respectively.
The results sparked fundamental and applied questions involving the use of AuNPs for the delivery of biomolecules to plants and elucidation of the currently elusive structure-function relationships of nanomaterials in plants. The work also elucidated the promise nanomaterials hold in harnessing plants for healthy crops, cheaper medicines, and more efficient biofuels. In addition, the project I conceived through my NSF GRFP proposal ultimately resulted in a Nature Nanotechnology publication. This experience further emboldened my career aspirations by 1) giving me the freedom to independently conceptualize and carry out a project while managing several failures along the way and 2) allowing me to lay a foundation for lab members to carry the work forward.
- Zhang, H.* Goh, N.S.*, Wang, J., Demirer, G.S., Butrus, S., Park, S-J, Landry, M.P. Nanoparticle Cellular Internalization is Not Required for RNA Delivery to Mature Plant Leaves. Nature Nanotechnology (2021) 17, 2: pp. 197-205.
- Wang, J.W., Grandio, E.G., Newkirk, G.M., Demirer, G.S., Butrus, S., Giraldo, J.P., Landry, M.P. Nanoparticle-mediated genetic engineering of plants. Molecular Plant (2019) 12; pp. 1037-1040.
- Butrus, S., Demirer, G., Goh, N., Zhang, H., Cunningham, F., Landry, M. Development and Characterization of Gold Nanoparticles for Plant Genetic Engineering
- Oral: UC Berkeley Amgen Scholars Program Symposium, Berkeley, CA (2018); ABRCMS Annual Meeting, Indianapolis, IN (2018); GCURS Rice University, Houston, TX (2018)
- Poster: UC Berkeley Amgen Scholars Program Symposium, Berkeley, CA (2018); BMES Annual Meeting, Atlanta, GA (2018); AIChE Annual Meeting, Pittsburgh, PA (2018)
This work was performed under the mentorship of Prof. Markita Landry at UC Berkeley within the Amgen Scholars Program.
II. Tuning size and charge of a multivalent polymer library for enhanced drug delivery to cartilage
Efficient transport of therapeutics through dense biological tissues has proven to be a complex challenge, especially for osteoarthritis (OA), a degenerative joint disease characterized by gradual cartilage degradation affecting ~30 million individuals annually. In this work, I focused on addressing the transport of a potential drug delivery vehicle into cartilage for the treatment of OA. The avascular, densely packed, and negatively charged extracellular matrix of cartilage has been a barrier to the transport and, thus, therapeutic effect of even locally administered drugs. I probed whether the uptake-to-cytotoxicity ratio of a nanocarrier in cartilage can be maximized by systematically tuning its physicochemical properties. Taking advantage of the negatively-charged cartilage, I tuned the size and charge of cationic polymers by grafting their periphery with a library of polyethylene glycol molecules. The results revealed a significant influence of size and charge on uptake capabilities of the polymers and their cytotoxicity in bovine cartilage explants and chondrocytes. Furthermore, these findings will inform key design principles of drug delivery vehicles for the potential treatment of OA patients.
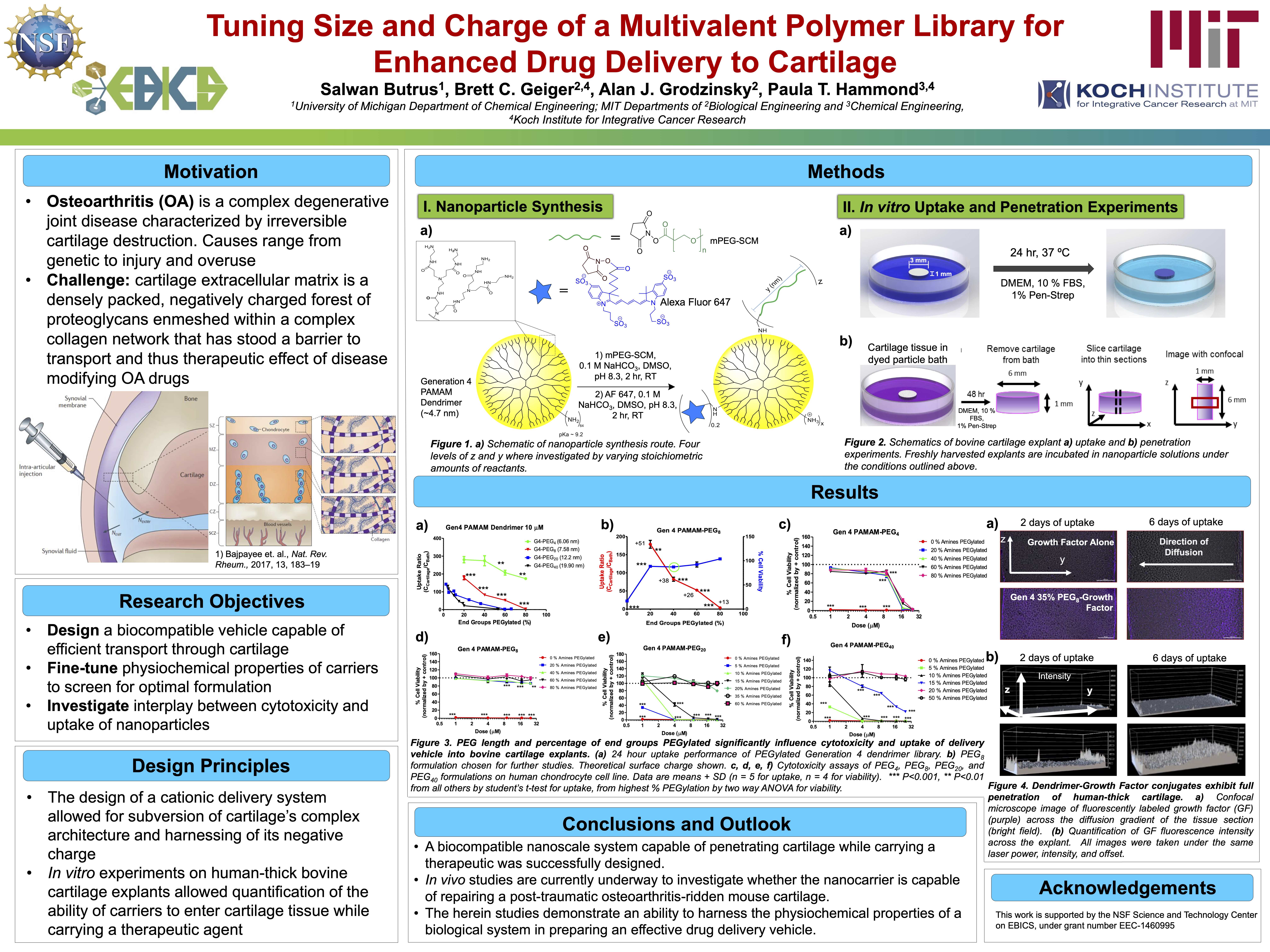
- Butrus, S., Geiger, B., Grodzinsky, A.J., Hammond, P.T., Tuning Size and Charge of a Multivalent Polymer Library for Enhanced Drug Delivery to Cartilage
- Oral: AIChE Annual Conference, Minneapolis, MN (2017)
- Poster: MIT Summer Research Programs Annual Poster Session, Cambridge, MA (2017); EBICS Annual Conference, Atlanta, GA (2017); BMES Annual Conference, Phoenix, AZ (2017); NanoDDS Annual Meeting, Ann Arbor, MI (2017)
This work was performed under the mentorship of Dr. Brett Geiger in Prof. Paula Hammond’s group at MIT as part of the NSF Center on EBICS.
III. Nanomechanical properties of zwitterionic polymer brushes
I began this project because I was intrigued by interfacial phenomena, namely those governing the performance of polymer brush coatings in supporting exogenous donor-free culture of human embryonic stem cells (hESCs). Prior work had indicated that surface rigidity and elasticity influence cell attachment, spreading, proliferation, and differentiation, but no studies had directly investigated the interaction between thickness, wettability, and nanomechanical properties of the surfaces developed by our group. I probed 1) whether simple reaction parameters that govern thickness and wettability can modulate the nanomechanical properties of these synthetic surfaces and 2) whether hESC propagation responded to these variations in architectural regimes.
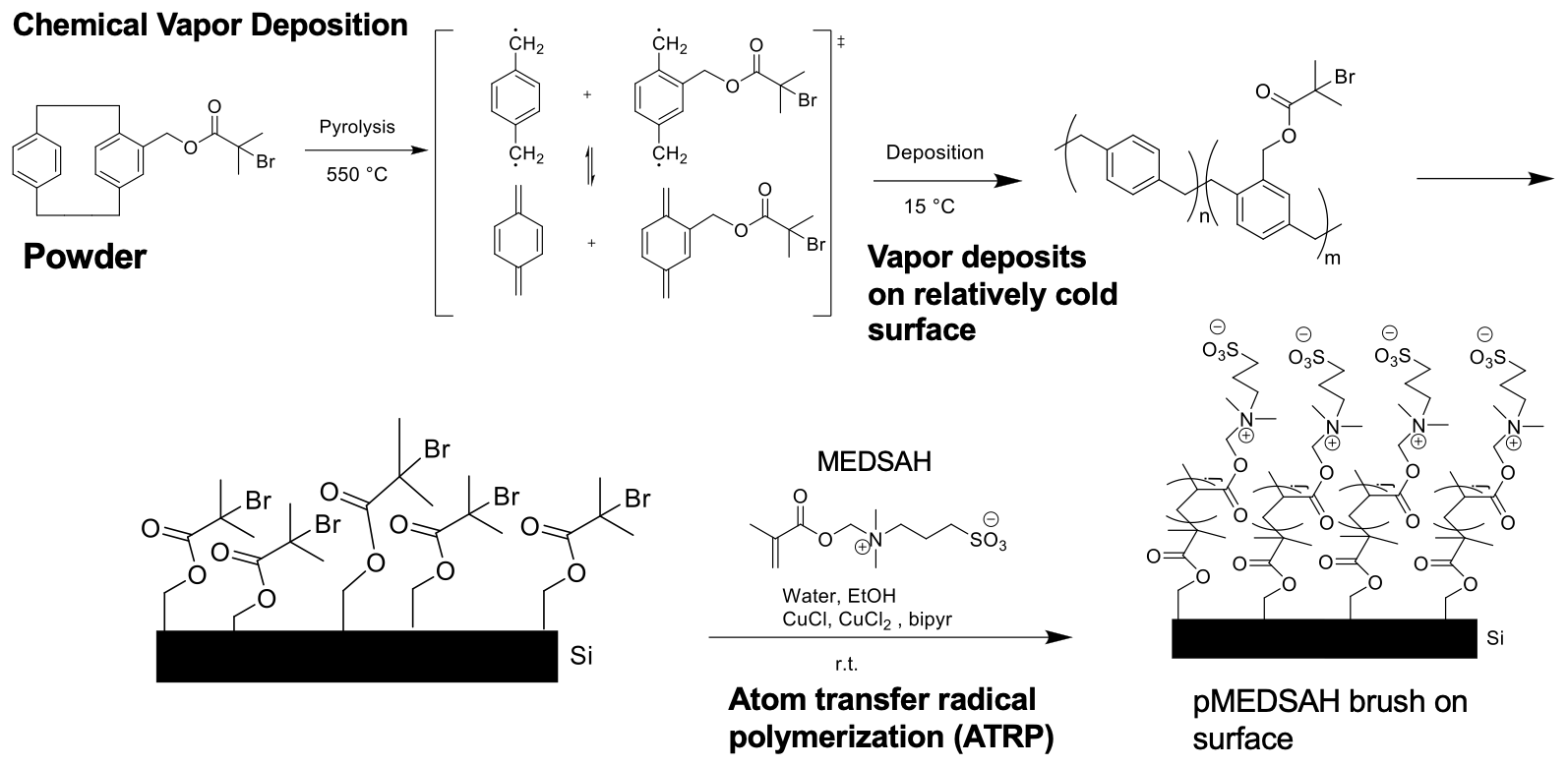
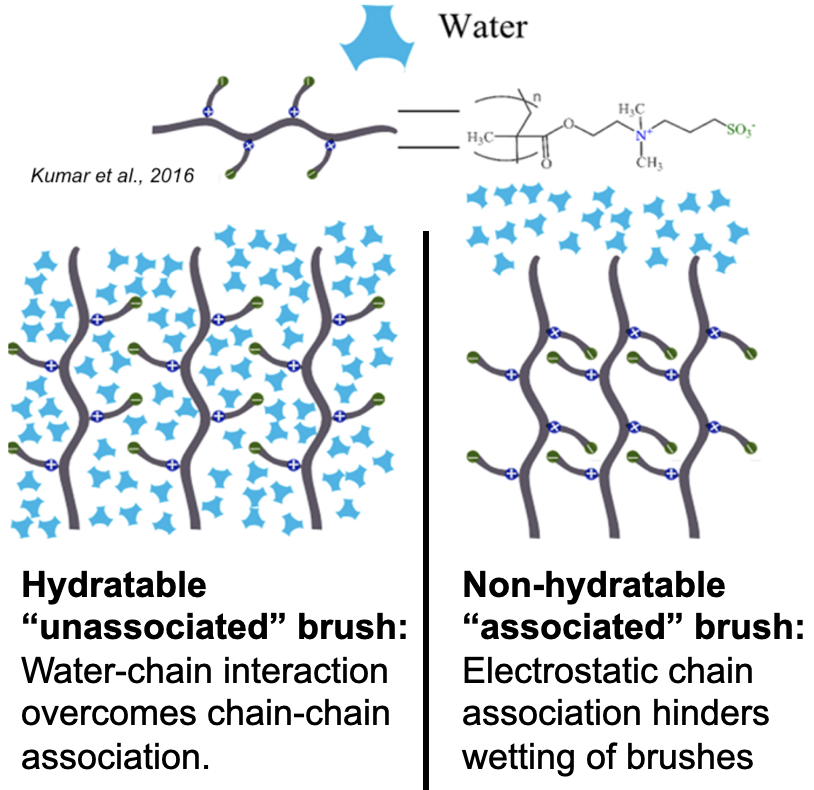
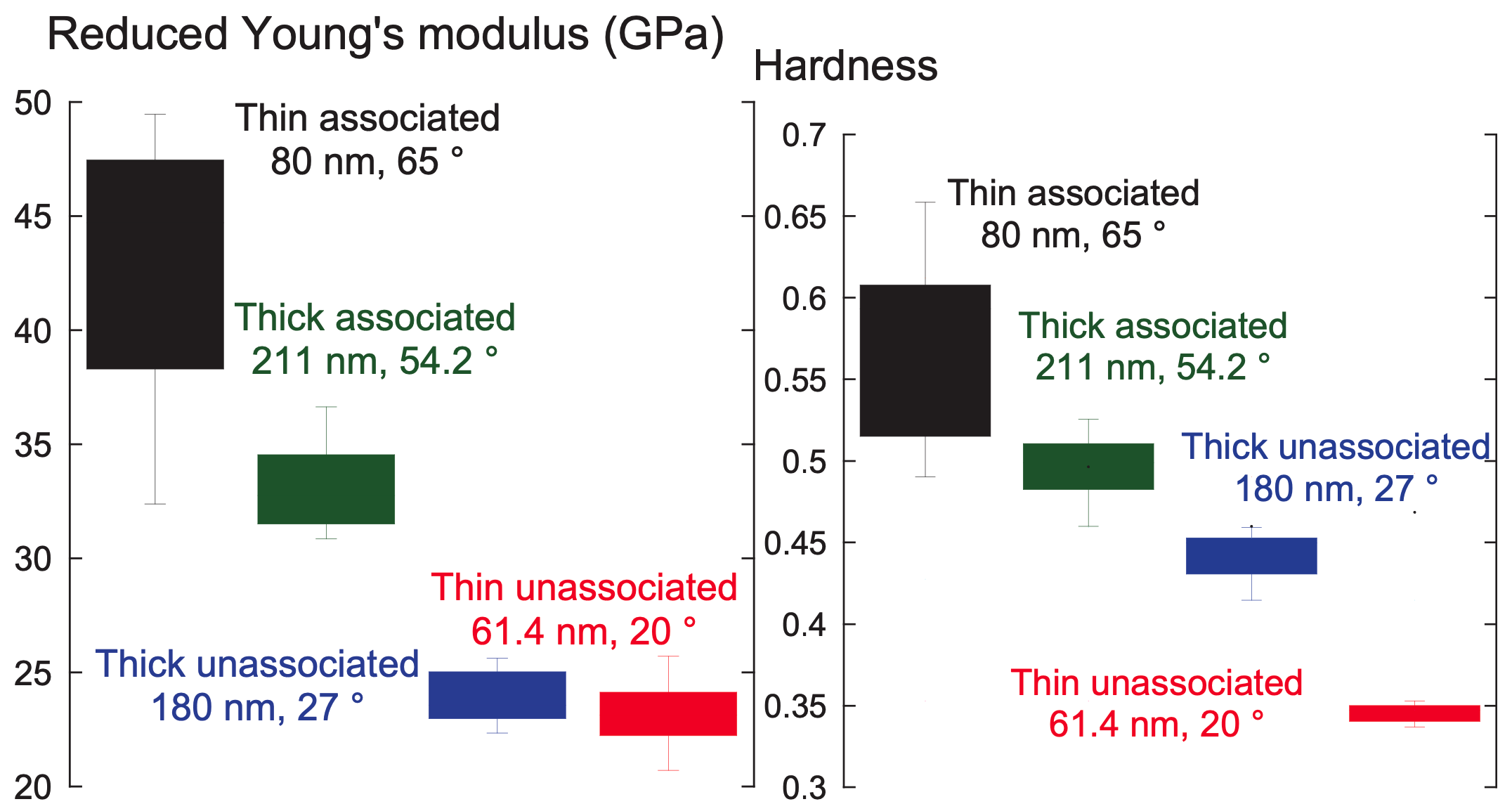
Through a statistical design of experiments approach, I employed chemical vapor deposition and atom transfer radical polymerization to design polymer brush coatings of varying thicknesses and wettabilities then acquired their reduced Young’s modulus, $E_r$, and hardness, $H$, via nanoindentation. Analysis of variance for $E_r$ and $H$ suggested that the nanomechanical properties are shaped by a complex interplay between thickness and wettability. This work explained previous results2 from our group, where cells preferred the moderately thick and moderately hydrophobic coatings because they had the optimal $ E_r $ and $H$ for rapid hESC propagation.
This work was performed under the mentorship of Prof. Ramya Kumar in Prof. Joerg Lahann’s group at the University of Michigan.